How is my search supported by taxonomies?
Last updated on December 13, 2023
Reaxys makes use of various taxonomies for tagging content and providing support for filtering search results in Quick search and in the Query builder.
Taxonomies are hierarchically organized controlled vocabularies. In Query builder they are used to enhance the Reaxys search experience in three ways:
- The hierarchical organization can give a quick overview of the content available for a particular data field
- The same organization makes it easy to search for closely related terms under a broader "umbrella" term or category.
- Taxonomy terms (concepts) typically are enriched with synonyms that enable the discovery of any terms with the same meaning that are in common use.
For instance, the authors of different publications may refer to the same target protein by different names, IDs, and abbreviations. The target taxonomy makes them discoverable under a unified concept with a harmonized display name, linked to all related synonyms.
To guarantee a high-quality standard, most of the taxonomies are fully manually curated.
For the medicinal chemistry domain, Reaxys uses taxonomies for the following data fields:
Field name | Update frequency |
|---|---|
Target Name | once per year |
Substance Effect | three times per year |
Substance Route of Adm. | three times per year |
Bioassay Animal Model | three times per year |
Biological Species | Target Species | three times per year |
(Clinical) findings/disease | three times per year |
Organs/Tissues | three times per year |
Cells/Cell Lines | three times per year |
Measurement Parameter | three times per year |
These taxonomies use data from internal and external authoritative sources.
From external sources:
Taxonomy | Source | Integrated external data |
|---|---|---|
Targets | UniProt (Swiss-Prot) | UniProt ID |
Targets | Protein Data Bank (via UniProt) - | PDB ID |
Targets | InterPro | InterPro ID |
Targets | Gene ontology | GO ID |
Cell lines | Cellosaurus - | Cellosaurus ID |
Species | NCBI Taxonomy - | NCBI taxid |
From internal sources:
Taxonomy data are harmonized to relevant Emtree branches.
Field name | Emtree branches |
|---|---|
Substance Route of Adm. | drug administration route |
Biological Species | Target Species | organisms |
Organs/Tissues | anatomical concepts |
(Clinical) findings /disease | diseases |
Bioassay Animal Model | experimental disease |
Cells/Cell Lines | cells |
Measurement Parameter | parameters |
Substance Effect | chemicals and drugs |
A quality workflow is in place for validating new concepts proposed during manual content excerption.
Targets and cell lines can be searched directly via Quick search using simple keywords or target names and identifiers as query. The other RTB taxonomies, as well as hierarchy browsers for targets and cell lines, are available from Query builder.
Figure 1 illustrates the use of RTB taxonomies for search via Query builder using the example of Substance Effect.
- Select 'Substance Effect' querylet from Target and Bioactivity section from right-hand bar. Open the taxonomy look-up by clicking this icon
 .
. - The taxonomy look-up allows browsing the hierarchy by expanding branches or searching for a specific concept by typing in the search box.
- Once the selection has been made, it can be transferred to the querylet by clicking on 'Transfer'.
- When searching in Substances, bioactivity data with the selected substance effect are shown under Bioactivity (Hit Data).
- The Heatmap view provides an overview on structure activity relationship by displaying by default chemical substances (Y-axis) vs biological targets (X-axis) and activity potency as pX (Cells).
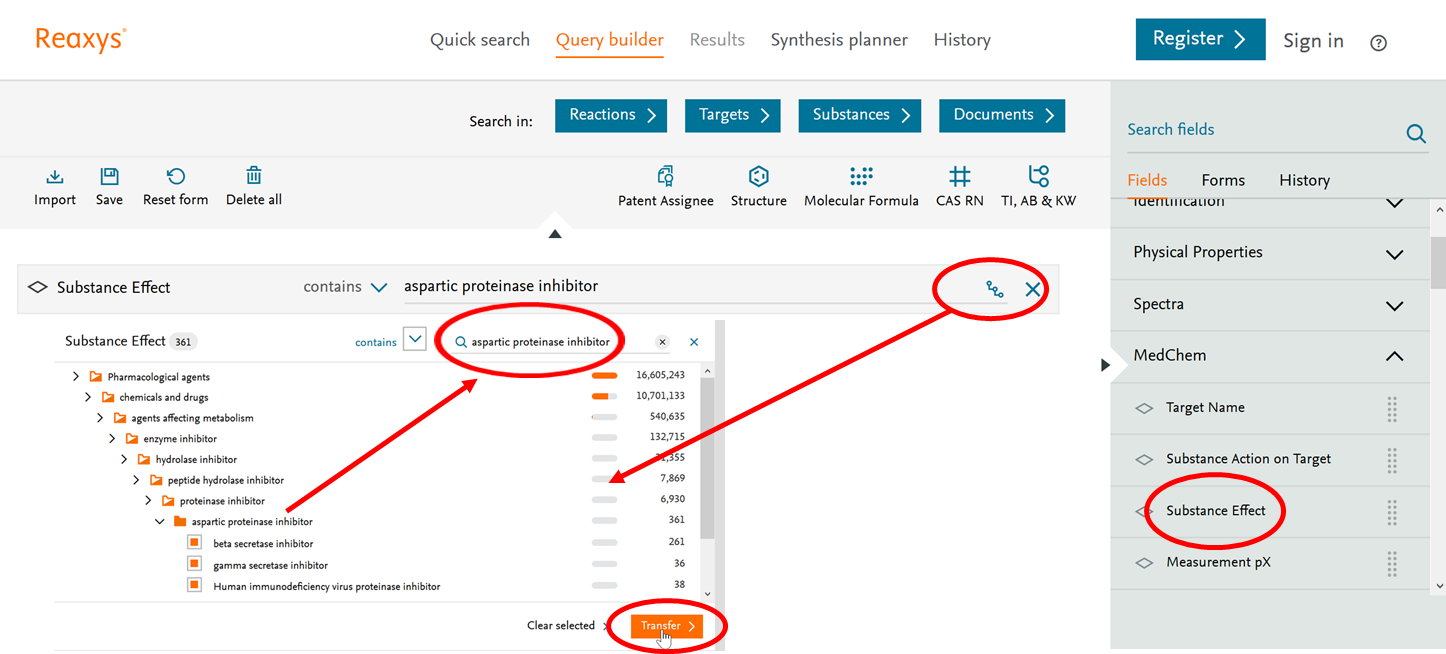 Figure 1: Query builder search
Figure 1: Query builder search
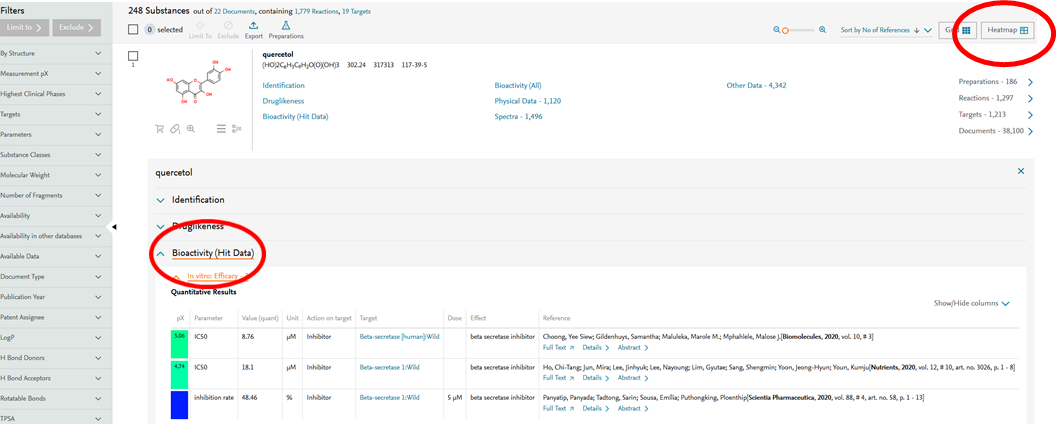 Figure 2: Results for search showing Substances
Figure 2: Results for search showing Substances
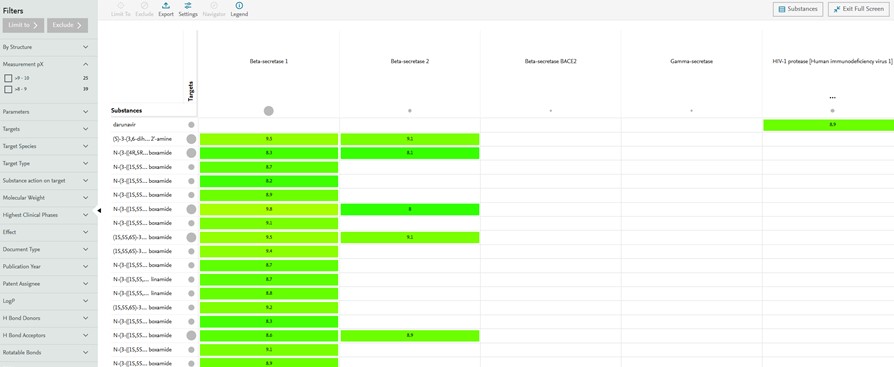 Figure 3: Bioactivity and Visualization
Figure 3: Bioactivity and Visualization
Executing a Quick search taxonomies are employed as available. Here is how to view or edit the synonyms that are included in your search on the example of the target taxonomy:
- Click 'Quick search' and type 'HDAC11' in the Search Reaxys field.
- Click on 'Find'.
- Results are available for Targets, Substances and Documents.
Clicking on 'View Results' for targets or substances leads to a search in quantitative bioassay data manually excerpted from literature, while the document search includes synonyms found in titles, abstracts, patent claims, or index terms originating from an automated annotation pipeline.
The Quick Search algorithm uses the target taxonomy to enhance and support the search experience by enriching an identified target name with known synonyms (“HDAC11” synonym of “histone deacetylase 11”). The result will also contain and expose links to external authoritative sources (e.g. UniProt)
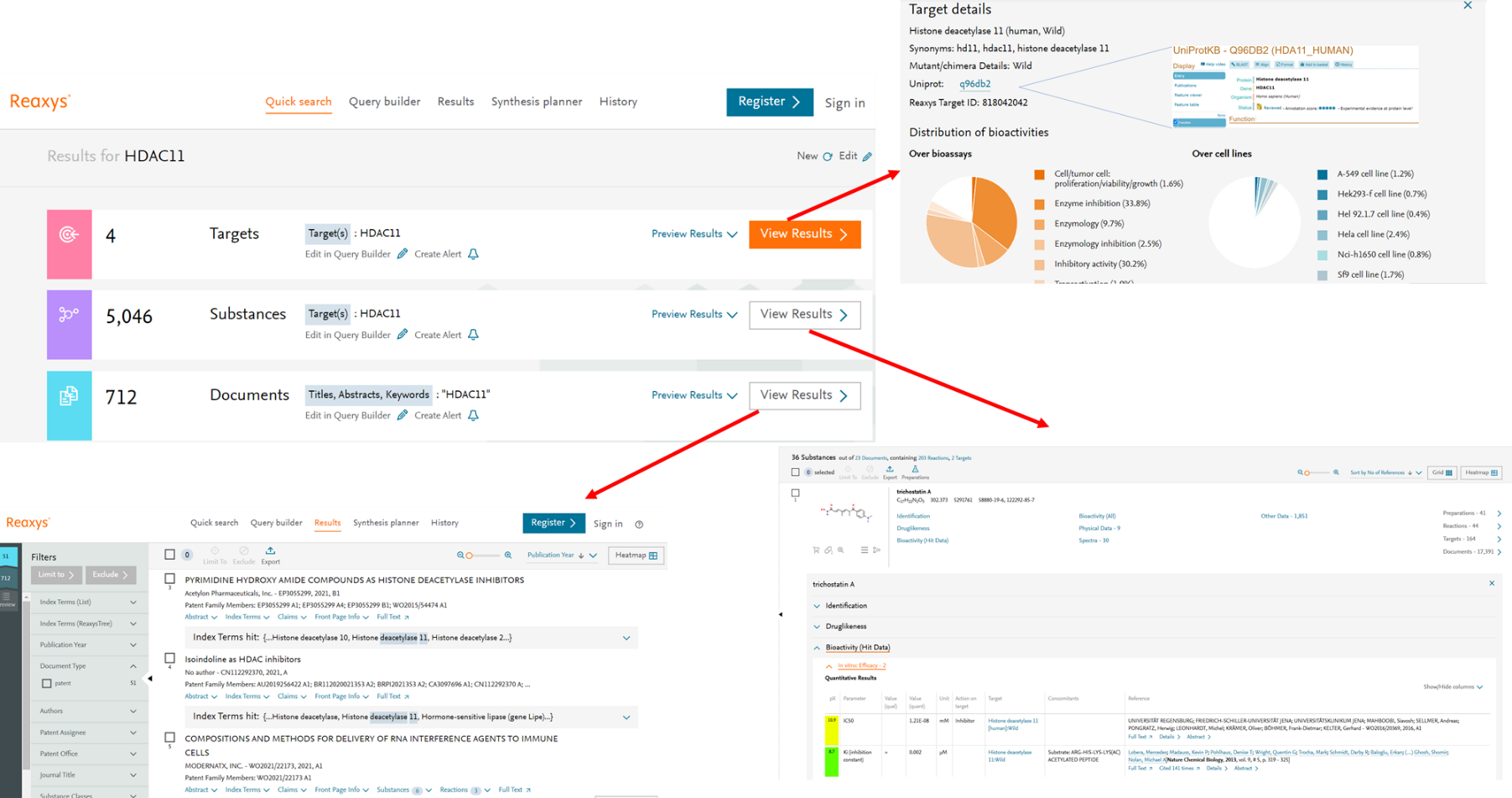
Figure 4
If you would like to view the taxonomy hierarchy or edit synonyms being used in a search, click on the Results page 'Edit in Query Builder'.
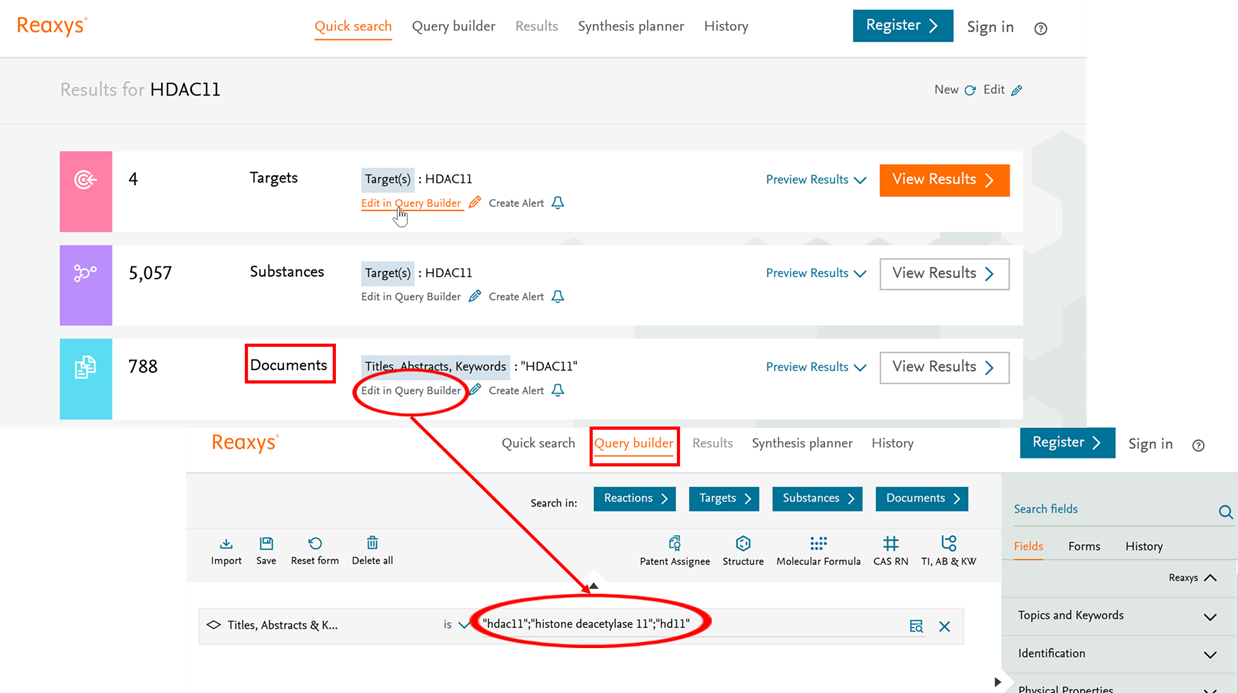
Figure 5
The target taxonomy combines data from external sources (Gene Ontology, InterPro, and UniProt) with a manually curated top level.
In addition, validated candidate terms from manual literature excerption are algorithmically added to the right place in the hierarchy. Species-unspecific nodes for grouping homologous targets from different species are added algorithmically as well. Figure 6 shows the schematic representation of the target taxonomy.
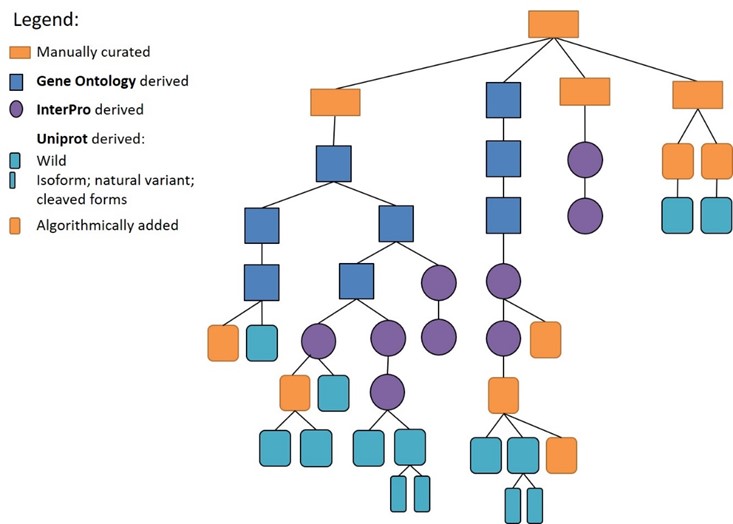
Figure 6: Schematic representation of how the different sources are built into the hierarchy of the target taxonomy
The target taxonomy contains information on genes and gene products (proteins/peptides), their alternative and short names and also contains the following external database identifiers: GO ID, InterPro ID, UniProt ID, PDB ID and NCBI taxonomy ID, when applicable. An example for a specific protein is shown in Figure 7. Please note that the full target taxonomy is trimmed on the user interface (UI) to show only the records that have data associated, the remainder of the records in the taxonomy are there and will be made visible to the user as soon as these are excerpted from literature.
Figure 7: Schematic representation of one of the two upward hierarchies for gene: MAPK3, Mitogen-activated protein kinase 3 [human]. * Please note the bottom layer contains the isoforms and natural variants of the wild protein for the UniProt record P27361 (currently not visible on Reaxys.com)
Did we answer your question?
Related answers
Recently viewed answers
Functionality disabled due to your cookie preferences