How do I create a batch query?
Last updated on October 16, 2025
Reaxys supports the execution of several queries in a batch of up to 1000 queries per batch. The searches are imported in Reaxys in either SD (collection of several molfiles) or textfiles (one advanced query per line).
You can use batch querying for structure searches (exact and sub-structure) and field searches in all contexts of the database. Reaxys searches batch queries sequentially and at the end presents the result combined into one single hitset. A report is available on the result page showing the results for each individual query in the batch. The report is available in the corresponding breadcrumb (view a screenshot in step 3).
How
Follow these steps to create a batch query in Reaxys:
Prepare your batch file in SD or Text file format:
SD file
- An SD File contains a text representation of the atoms, bonds, connectivity and coordinates of one or more structures.
- Each individual structure is called a molfile.
- Molfiles are separated by "$$$$" in an SD File.
- SD Files are widely used in cheminformatics and are created and read by many different applications.
- The SD file needs to have an .sdf extension in order for Reaxys to recognize the file format.
------- start copying the text from next line ------
HDR
10 10 0 0 0 0 0 0 0 0999 V2000
1.4995 0.8652 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0000 1.7315 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4995 0.8652 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0000 0.0000 0.0000 O 0 0 0 0 0 0 0 0 0 0 0 0
1.4995 2.5978 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
0.0000 1.7315 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
3.0001 1.7315 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
2.4995 2.5978 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1.0000 3.4641 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
4.0001 1.7315 0.0000 C 0 0 0 0 0 0 0 0 0 0 0 0
1 2 1 0 0 0 0
1 3 1 0 0 0 0
1 4 1 0 0 0 0
2 5 1 0 0 0 0
2 6 1 0 0 0 0
3 7 1 0 0 0 0
5 8 1 0 0 0 0
5 9 1 0 0 0 0
7 8 1 0 0 0 0
7 10 1 0 0 0 0
M REG 791935
M END
> <description>
Derivatives of cyclohexanol
> <name>
2,3,5-trimethylcyclohexanol
$$$$
------- stop copying the text with the line above, paste it into a text editor and save it as a file with the extension ".sdf" ----------
Text file
- A simple text document (structured with Carriage return/Line feed) containing single-byte characters.
- Each line of the text file is interpreted as one query.
- Structures can be embedded as SMILES (line notation for molecules using letters and other characters) or molfiles (See SD File).
- The text file may contain several SMILES or MOLFILES, but only one is allowed per line/query.
- The Text file needs to have a .txt extension, otherwise it won’t be recognized by Reaxys.
Example 1: Create a text file for searching identifiers (here: a list of patent numbers.)
Open your text editor, then
------- start copying the text from next line ------
CIT.PN=US2009/8002
CIT.PN=US2009/8605
CIT.PN=US2009/10846
CIT.PN=US2009/10865
CIT.PN=US2009/10870
CIT.PN=US2009/10884
------- stop copying the text with the line above, paste it into a text editor and save it as a file with the extension ".txt" ----------
Example 2: Create a text file with a combined property and structure query (here: search for 1,4-derivatives of 2-chloro-resorcinol as a molfile with melting point 80-100):
------- start copying the text from next line ------
MP.MP=80-100 AND MOLFILE='\n\nHDR\n 0 0 0 0 0 0 0 0 0 0999 V3000\nM V30 BEGIN CTAB\nM V30 COUNTS 9 9 0 0 0\nM V30 BEGIN ATOM\nM V30 1 C 5.9701 5.7801 0.0000 0\nM V30 2 C 5.9701 4.7681 0.0000 0\nM V30 3 C 5.0941 4.2701 0.0000 0\nM V30 4 C 4.2361 4.7681 0.0000 0\nM V30 5 C 4.2361 5.7801 0.0000 0 SUBST=6\nM V30 6 C 5.0941 6.2781 0.0000 0\nM V30 7 O 5.0941 7.2722 0.0000 0\nM V30 8 Cl 6.8272 6.2781 0.0000 0\nM V30 9 O 6.8272 4.2701 0.0000 0 SUBST=6\nM V30 END ATOM\nM V30 BEGIN BOND\nM V30 1 2 1 2\nM V30 2 1 1 6\nM V30 3 1 1 8\nM V30 4 1 2 3\nM V30 5 1 2 9\nM V30 6 2 3 4\nM V30 7 1 4 5\nM V30 8 2 5 6\nM V30 9 1 6 7\nM V30 END BOND\nM V30 END CTAB\nM END\n'
------- stop copying the text a line above, paste it into a text editor and save it as a file with the extension ".txt" ----------
Example 3: Create a text file for searching SMILES strings
Open your text editor, then
------- start copying the text from next line ------
SMILES='CC#CC1CCCC1'
SMILES='N1C=CC2=C1C=CC=C2'
SMILES='CC(C(C1=CC=CC=C1)O)NC'
------- stop copying the text with the line above, paste it into a text editor and save it as a file with the extension ".txt" ----------
- Click ‘Import’ on the left side of the top banner on the Quick Search (view screenshot) or Query Builder page (view screenshot).
- Browse for your file or drag and drop it in the highlighted field.
- Click ‘Search’ in the top banner and select the result type (i.e. reactions, documents, targets or substances) from the dropdown.
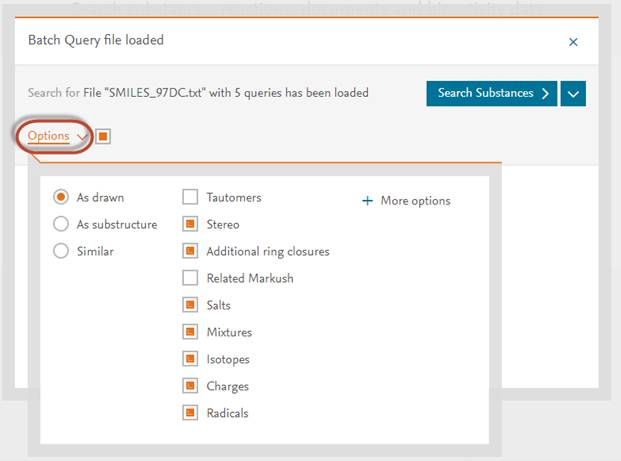
Note: To change the structure editor preferences, such as substructure search, please click on ‘Options’ as in screenshot below:
- The Results page will open displaying each resulting hit (the same way results from a non-batch query would be displayed). However, if the same hit correlates to more than one structure in the query it will appear only once in the results display.
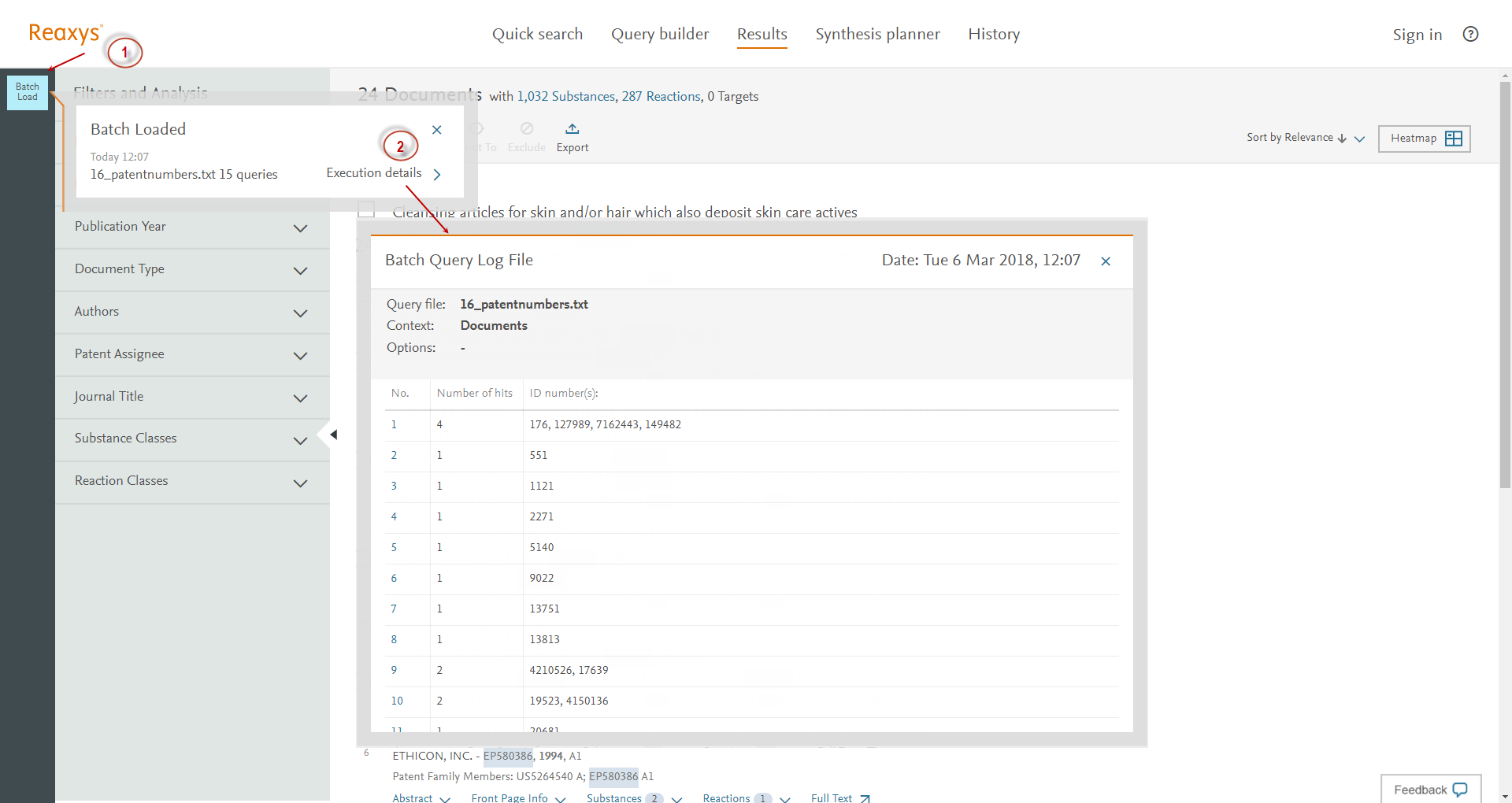
A batch query generates a log file with an overview of the search. Click the 'Batch query' breadcrumb (1) at the top left side of your screen to see the progress and information on your batch query. To see more detailed information, click 'Execution Details' (2).
Did we answer your question?
Related answers
Recently viewed answers
Functionality disabled due to your cookie preferences