How is the Substance Similarity Score calculated?
Last updated on November 18, 2025
The Substance Similarity Score is calculated using a proprietary algorithm. It is based on a Tanimoto-type similarity comparison between structural feature vectors derived from both the query structure and each candidate in the database. The vectors are constructed from 512 commonly occurring chain and ring fragments in Reaxys (e.g., C=C, C=C–C, etc.).
Each substance is represented by a property vector (p1) that encodes these key structural features. This property vector is then combined with additional molecular-formula–based descriptors (p2) to form the final similarity vector. The resulting vector representation allows the system to efficiently and accurately assess molecular similarity within the Reaxys platform.
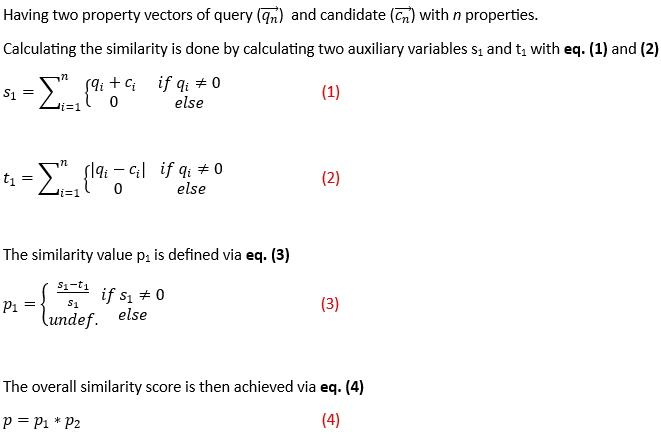
Using these property (p1) and molecular-formula vectors (p2), the similarity score is calculated as described below.
Description of the algorithm to calculate the similarity value, illustrated here for the property-vector component (p1) of the similarity score:
Did we answer your question?
Recently viewed answers
Functionality disabled due to your cookie preferences